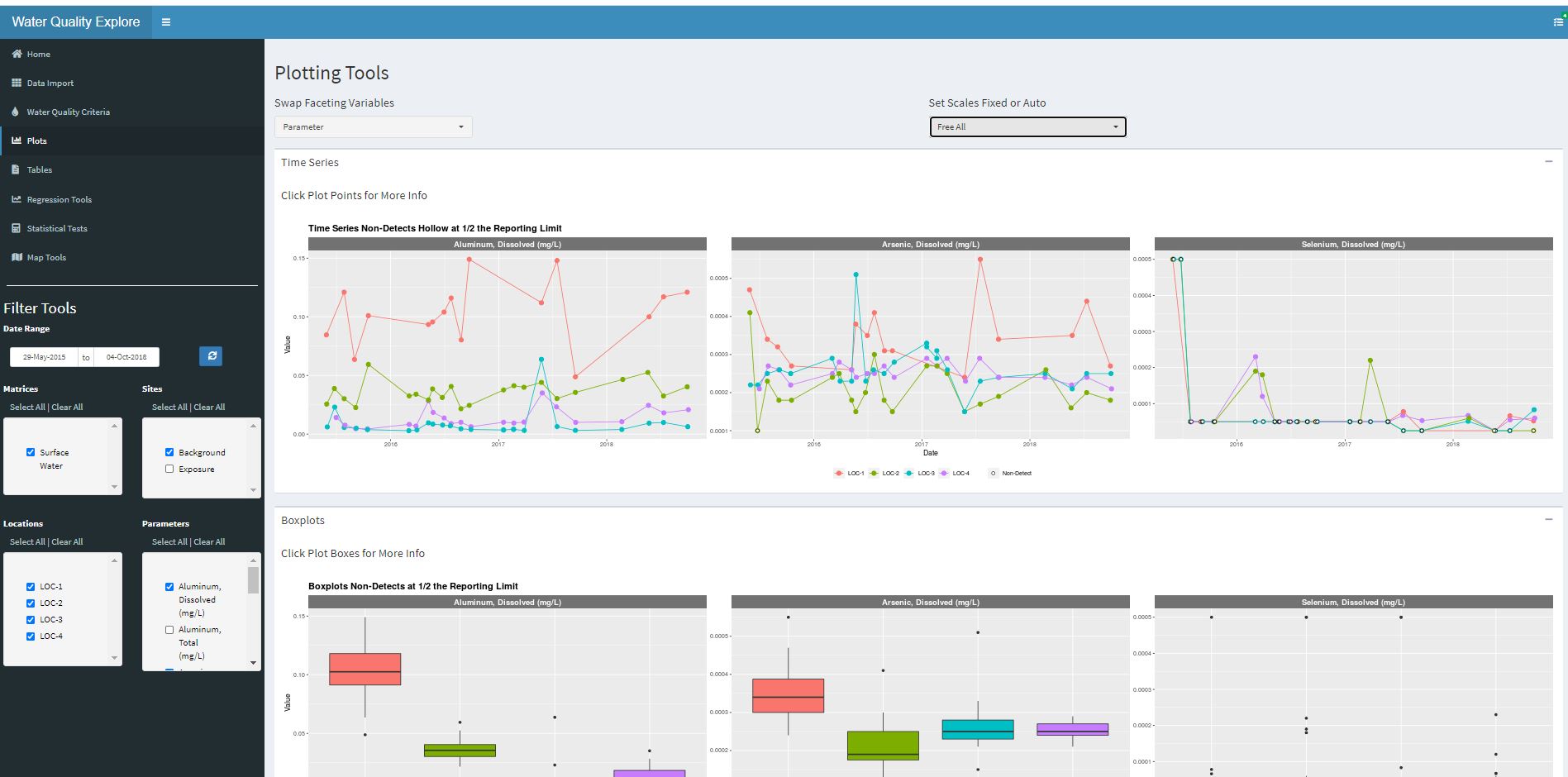
Example Data Products
Data Products
Web Applications with Shiny
Try my R Shiny App for exploring environmental datasets. Follow the link, hit the “Load Demo Data” button, pick a few locations and analytes from the left side, and explore the demo data.
Try my R Shiny app…
My Demo Data Explorer AppVisualizations
Any common analytic visualizations can be created as images or PDF files, for insertion into a presentation, a document or to put in an appendix.
Any kind of plots, too many to show. Histograms, scatter plots, bar charts, box plots, piper diagrams, stiff plots, heat maps, contour maps, bi plots, pies, control charts, radial charts, bubble….
Box or Violin Density Compares Variables between Groups
df<-iris %>%
gather(Parameter,Value,1:4)
ggplot(df,aes(Species,Value,fill=Species))+
geom_jitter(alpha=.5)+
geom_boxplot(alpha=.75)+
facet_wrap(~Parameter)+
theme(legend.position='bottom',legend.title=element_blank(),
strip.background = element_rect(fill='gray69'))+
labs(y="Dimension (cm)",title="Box Plots, Sepal and Petal Morphology\nin Three Iris Species")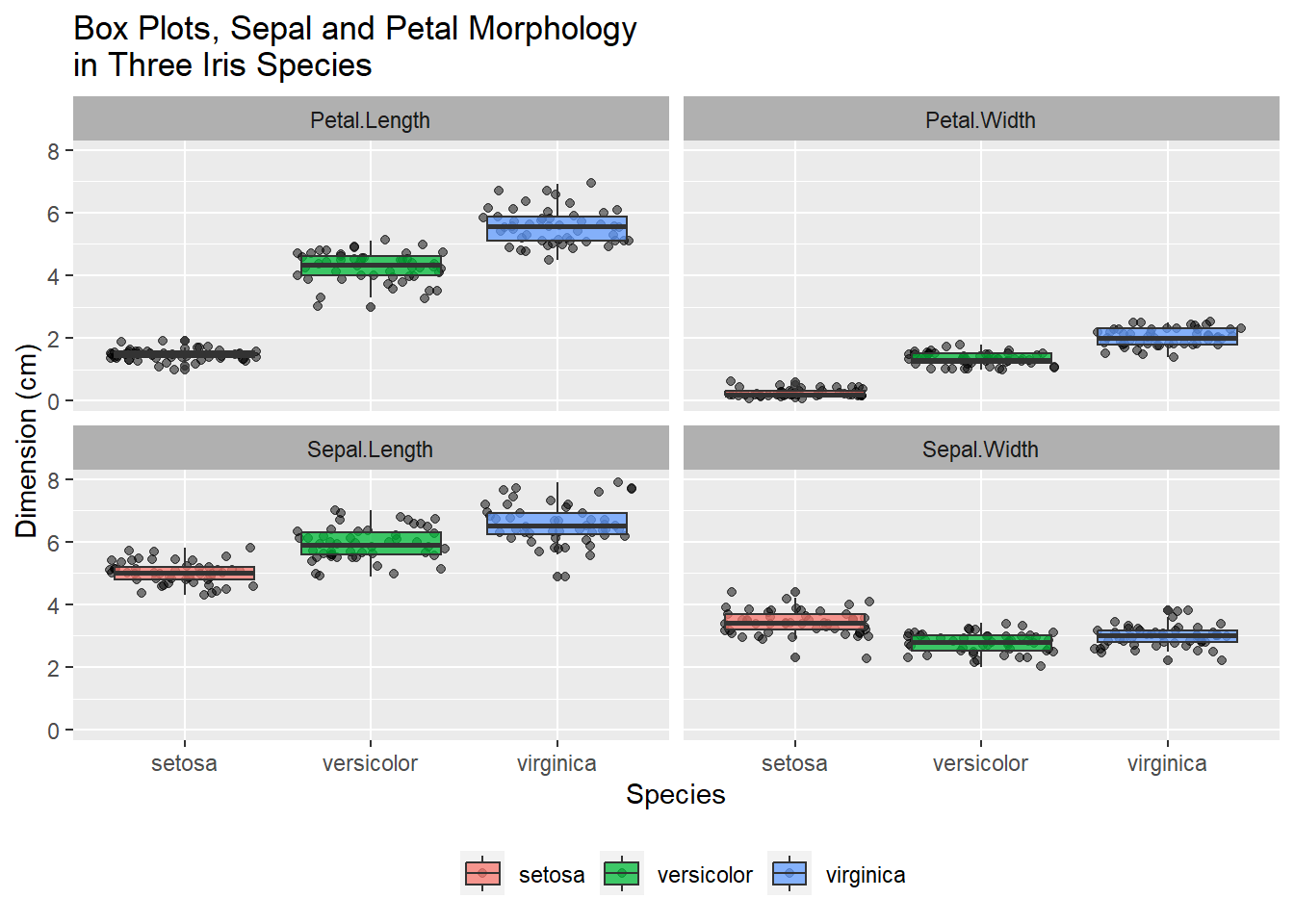
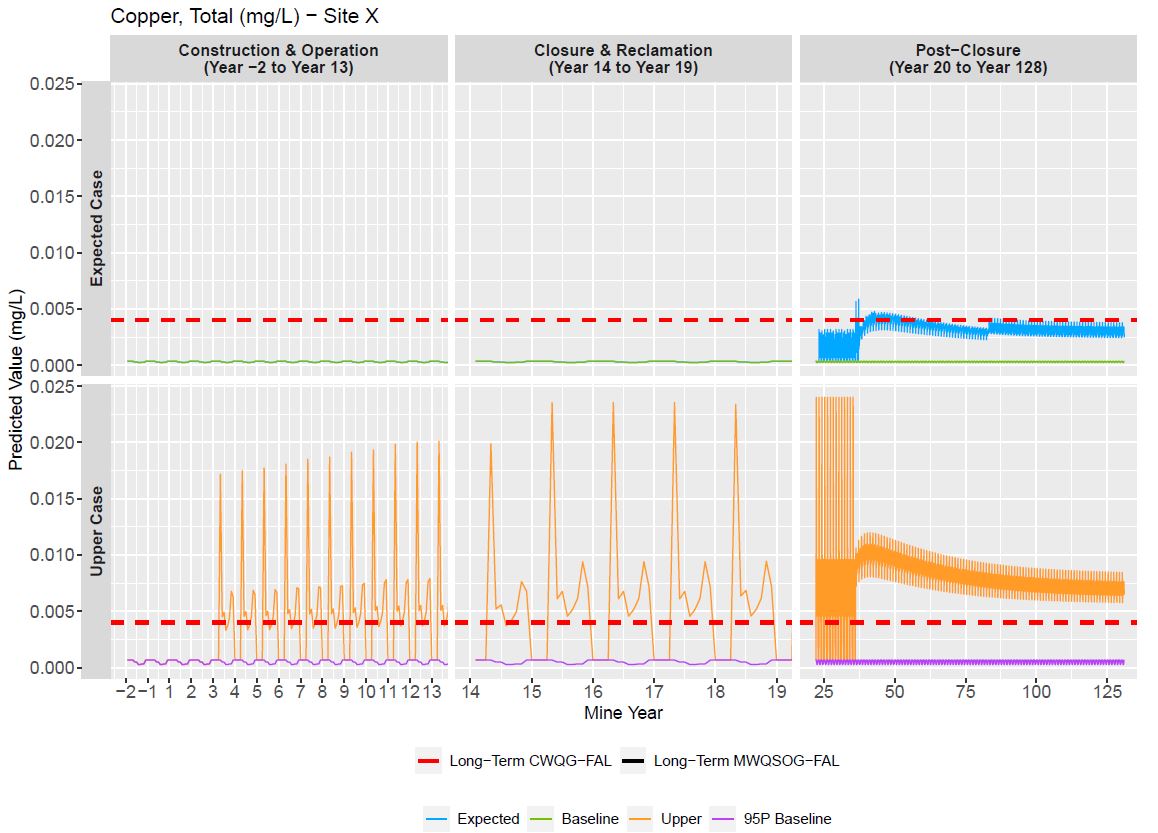
Time Series/Grouped Time Series Depict Change through Time
GoldSim model output for two modeling scenarios, over the
three expected phases of mine operation.
A look at molybdenum concentration behavior by
season. 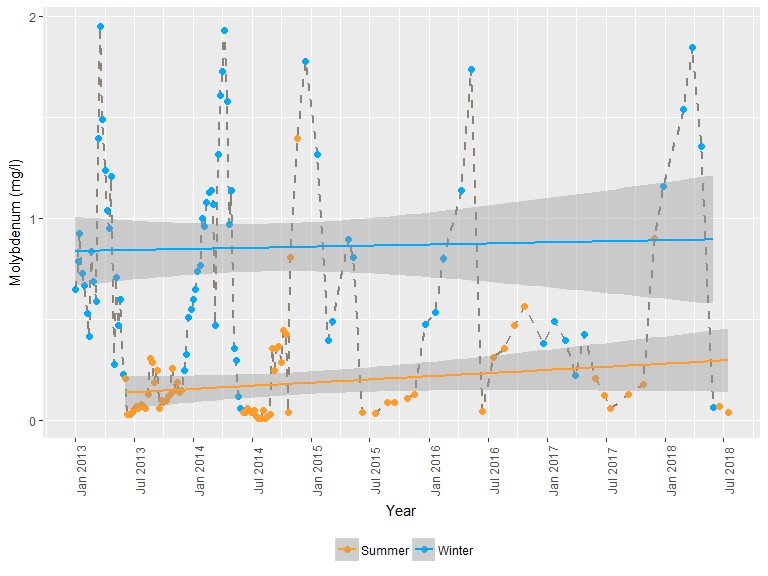
Regression reveals Correlation between two Variables
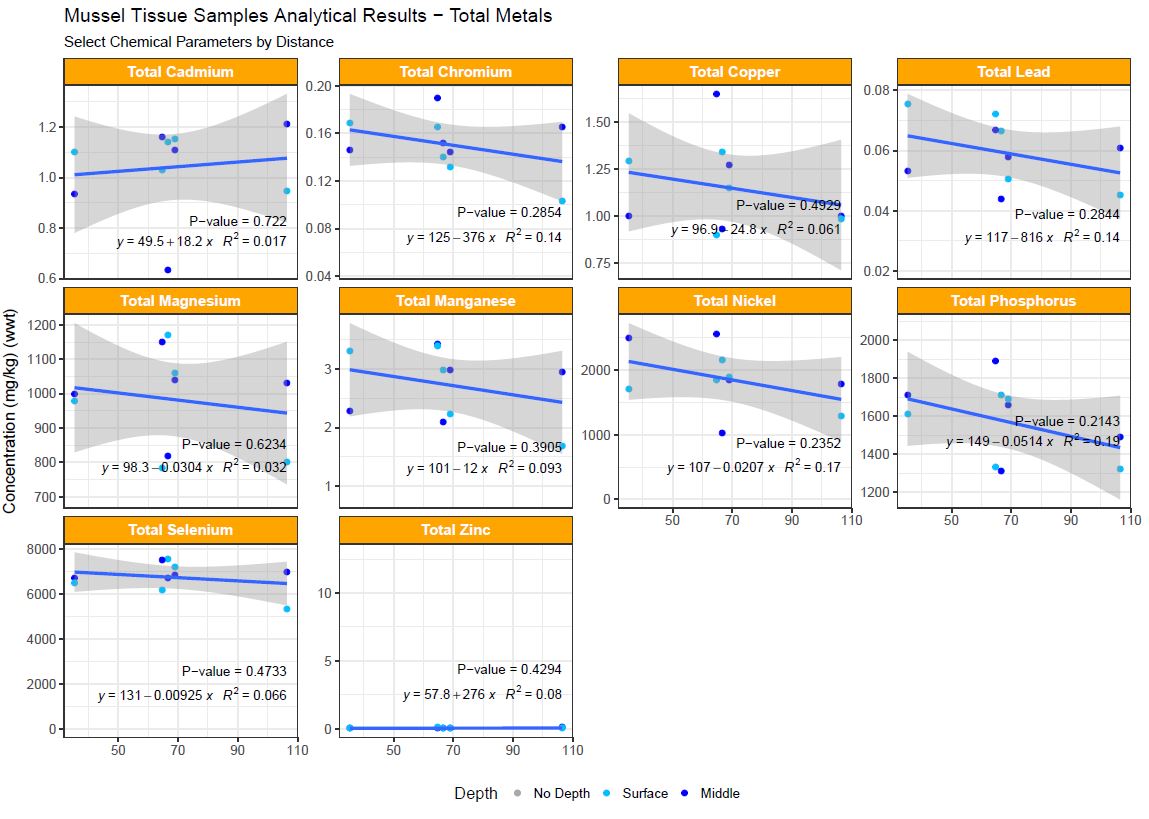
Marine mussels tissue contaminant as a function of distance from the source.
A look at arsenic’s correlation with several other
variables. 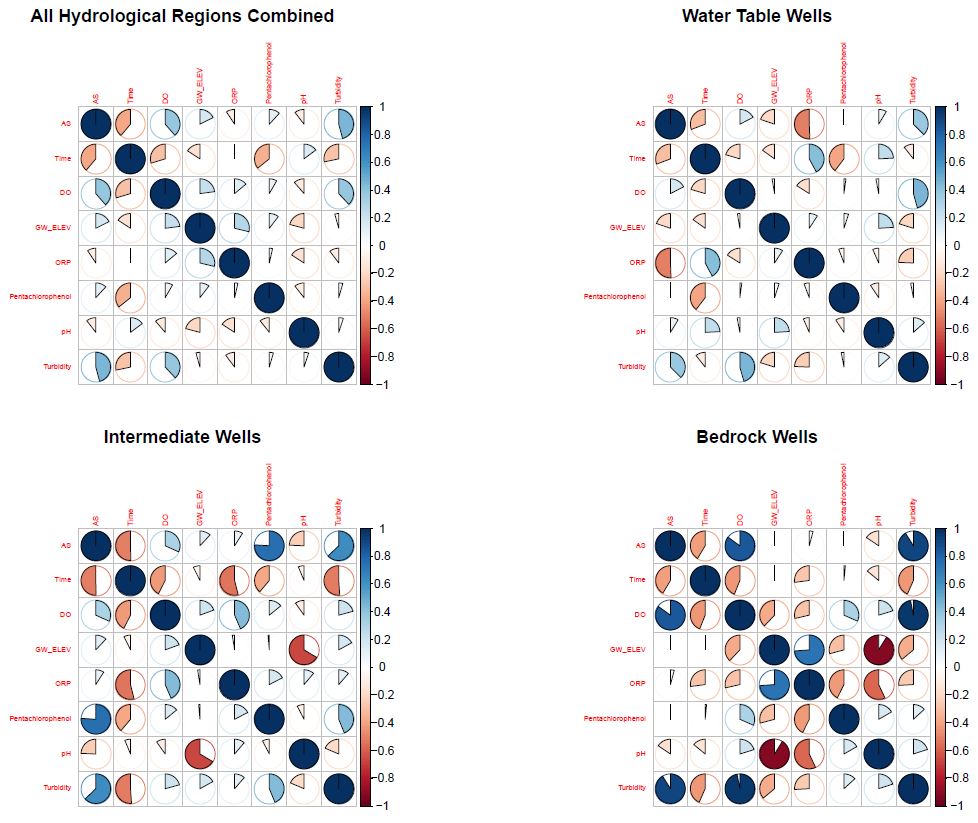
Interactive Visualizations
Interactive visualizations can be presented in PowerPoint, HTML files, or in HTML slide presentations, allowing the user or presenter to interact with the data in real time.
An Interactive Time Series of COVID-19 Data in the US
Hover the plot to activate the toolbar, filter the data by double-clicking in the legend to select a location,and using the Ctrl key + click to select multiple locations to compare.
conData<-fread('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_US.csv')
dData<-fread('https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_deaths_US.csv')
cdata<-conData %>%
select(7:ncol(.)) %>%
gather(Date,Result,8:ncol(.)) %>%
mutate(Parameter = 'Confirmed Cases')
ddata<-dData %>%
select(7:ncol(.)) %>%
gather(Date,Result,8:ncol(.)) %>%
mutate(Parameter = 'Deaths')
gdata<-bind_rows(cdata,ddata) %>%
mutate(Date = as.Date(Date,format='%m/%d/%y')) %>%
group_by(Parameter,Province_State,Date) %>%
summarize(Result = sum(Result)) %>%
arrange(Parameter,Province_State,Date) %>%
as.data.frame(.) %>%
filter(#Parameter=='Confirmed Cases',
Result>0 & Result != '')
n<-length(unique(gdata$Province_State))
g<-ggplot(gdata,aes(Date,Result,color=Province_State))+
theme(plot.margin = margin(0.5,0.1,0.1,0.1,unit = 'in'))+
geom_line()+
theme(legend.position='None')+
#scale_color_manual(values = rep('grey',n))+
facet_wrap(~Parameter,scales='free')+
labs(y='# of Cases',title = 'COVID-19 Cases through Time by State/Province')
ggplotly(g,width=800,height=500) %>%
layout(hoverlabel=list(bgcolor="red")) %>%
highlight(on = "plotly_hover")